Whether belonging to a rooster or an airplane, a wing is a biologically inspired structure, tested in nature and optimized by trial and error. To help qualify wing designs for ascension from one-off observations into practical, routine use, a checklist of well-defined analytics parameters to reliably harness their power may be necessary. Likewise, for investigators in the thriving field of extracellular vesicles (EVs) to earn their own flightworthy wings, a group of experts from the International Society of Extracellular Vesicles (ISEV) published a set of guidelines known as MISEV (Minimal Information for Studies of Extracellular Vesicles).
Here we’ll focus on the characterization guidelines from the most recently published version, MISEV 2018. 1 We intend this blog post to remind the reader about the existence of these guidelines, inform an understanding of their purpose, and provide a quick overview of the recommendations as they pertain to state-of-the-art analytics assays. We urge the reader to consult the MISEV publications directly to ensure they are producing work that may ultimately contribute to the collective body of EV research.
Extracellular Vesicles (sometimes called “exosomes”) are small membrane-bound particles, ranging from 30nm to several microns in size, 2 that are released by cells into the extracellular environment. Whether these are microvesicles (MVs) that bleb from the plasma membrane or exosomes that form internally out of multi-vesicular bodies, EVs contain various types of bioactive molecules. These “cargoes” include proteins, nucleic acids, metabolites, and lipids, and they’ve been found to play an important role in intercellular communication. 3
In November 2022, we published a blog post to summarize our recent webinar about EV Analytics and describe the modalities we use in-house to analyze and characterize the EVs we produce. We also discussed why standardized analytics work is important to develop EV-based therapeutics. 4 As most folks reading this already know, EVs are a potential source of new and exciting treatments. 5, 6 Since the EVs’ biomolecule cargoes can act as the bioactive components of these drugs, it is clear why the analyses of EV contents and surfaces are essential for identifying and understanding the mechanisms of these potential therapeutics, and for assessing drug potency across manufacturing lots.
But as this young field evolves, the definition and classification of EVs are moving targets. Beyond the use of EVs as drug products or drug delivery vehicles, EVs are also avidly studied to assess their potential as diagnostic and prognostic biomarkers, 7, 8, 9 to elucidate their influence in biological processes including immune responses, 10 tissue regeneration, 11 and cancer metastasis 12 —and much more.
MISEV guidelines are updated intermittently, with the first iteration published in 2014 13 and an update provided in 2018 1 that included feedback from a much larger group of ISEV scientists. Draft guidelines were released in 2022 but are yet to be officially published. 14 The purpose of these guidelines is to provide a framework for researchers to report their EV studies in a uniform and transparent manner, thereby facilitating data comparison and reproducibility across different laboratories and experimental conditions. They provide direction on five main areas of EV research: collection and pre-processing, separation and concentration, characterization, functional studies, and reporting and data interpretation.
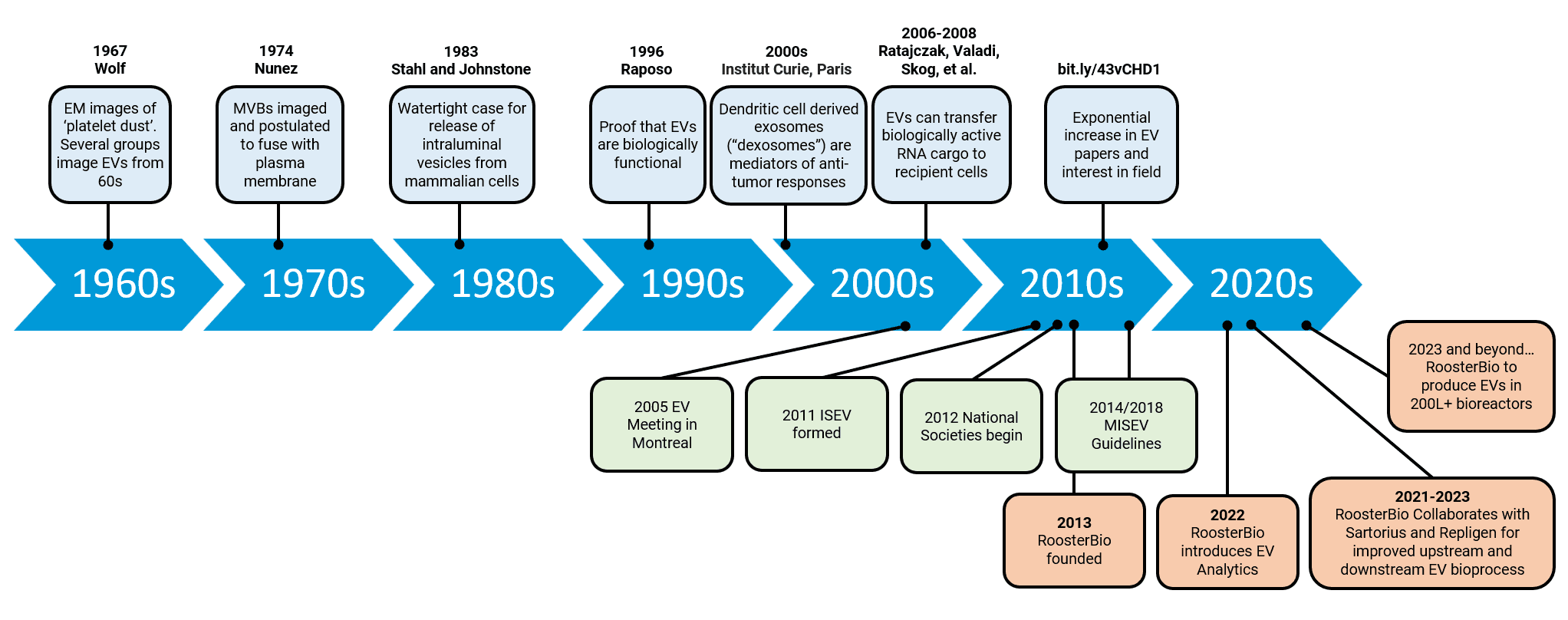
Figure 1. A history of EV research. The MISEV guidelines are a relatively recent introduction to the field and the more scientists work within these guidelines, the more comparable, consistent, and reproducible their research will be and the field can advance faster as a whole. Adapted from Couch 2020 et al. 15
It is broadly understood that the three main types of EVs are microvesicles, exosomes, and apoptotic bodies, and they are categorized by a range of characteristics including their size, from where in the cell they originated, how they are released, their content, and how they function. 16 However, the MISEV 2018 guidelines recommend they be generally classified by size with small EVs (sEVs) <200nm and medium/large EVs (m/lEVs) >200nm, and they discourage the use of such labels unless the cellular origin of the EV can be shown. 14 For this blog, we conventionally refer to small EVs which are currently thought to be most therapeutically relevant and most important to our customer base.
ISEV recommends the following information be available for each batch of EVs prepared:
- Quantity of the EV source (e.g. number of cells, volume of fluid, tissue mass).
- Quantity of EVs to the extent possible (e.g. total particle number and/or protein or lipid content).
- Characterization of markers or components associated with EVs or their subtypes (e.g. lipid bilayer, surface markers, cytosolic markers).
- Characterization of presence of non-vesicular, co-isolated components (e.g. albumin).
This information can be summarized according to MISEV’s checklist that helps govern the readiness of each experiment—used with EV material—for exposure in higher quality peer-reviewed publication or regulatory filings. RoosterBio has recently expanded its capabilities to provide standard analytical services that approach the MISEV checklist as depicted in Table 2a (Sections 4a, 4b and 4c) of the MISEV 2018 Position Statement (Théry, et al.). Below, we provide for your reference tabled information of item-by-item examples of ways we quantify (Section 4a), globally characterize pooled EV materials (Section 4b), and characterize single EVs (Section 4c). For EV quantification, we employ the Attune NxT (ThermoFisher), Biotek Synergy 2 (Agilent), NanoFCM Flow NanoAnalyzer (NanoFCM), NanoSight NS300 (NanoSight), NucleoCounter® NC-200™ (Chemometic), and ZetaView (Particle Metrix) instrument platforms. For EV characterization, we use the Agilent Bioanalyzer 2100 (Agilent), Biotek Synergy 2, ProteinSimple Jess (bio-techne), and QuantStudio 5 RT-PCR (ThermoFisher) systems. To characterize single-particle EV data events read from a population-wide sample, we work with the multi-parameter, molecular phenotyping features of the NanoFCM in concert with corroborating particle size data from the NanoSight NTA.
Our first table (below) outlines various instrumentation and assays used for the quantification of EVs in concert with MISEV Checklist Section 4a. Each entry includes the importance level, data to capture, instrumentation name, instrument type, and the specific assay being used. The importance level for all entries is classified by MISEV as “mandatory,” and involves quantification of originating cell material as well as the bioproduced EVs/exosomes.
Data to capture on quantified EV material is ideally from two corroborating methods, expressed per volume of initial fluid and/or number of producing cells/mass of tissue by a cell counter (NucleoCounter) and/or a flow cytometer (Attune Nxt). Factoring into that, we gather data points of EV material protein amount (BCA assay for absorbance on a Biotek Syn2 plate reader) and/or counted EV particle number (NanoSight), and/or EV lipid amount with the ZetaView.
MISEV 2018 Section 4: EV Characterization Checklist
+++Mandatory / ++ Mandatory if applicable / + Encouraged
| 1. Quantification (Section 4-a, Table 2a) | ||||
| Importance | Description | RoosterBio Instrumentation | Rooster Assay | |
| +++ Mandatory | Volume of fluid, and/or cell number, and/or tissue mass used to isolate EVs |  |
NucleoCounter 17
Cell counter – count viable cells in suspension |
Cell counts |
 |
Attune NxT
Flow Cytometer –understand cell type via surface markers |
Custom assay | ||
| +++ Mandatory | Global quantification by at least 2 methods, expressed per volume of initial fluid and/or number of producing cells/mass of tissue:
|
 |
NanoSight 18
Nano Tracking Analysis – assess particle count and size distribution |
EV Concentration in particles/mL and EV diameter distribution |
 |
ZetaView 19
Particle Counter – counts, particle size, and purity with lipid staining |
Count and % of particle population with membrane-bound lipids | ||
 |
NanoFCM
Nanoparticle Flow Cytometry – counts, particle size, purity, surface markers and specific cargo detection |
Custom Assay | ||
 |
Biotek Syn2 20
Plate Reader – absorbance, luminescence, and fluorescence for protein quantification |
Lipid Quantification, Total Protein | ||
| +++ Mandatory
|
Ratio of the 2 quantification figures | NA | ||
Our next table (below) outlines various instrumentation and assays used for the global characterization of EVs in concert with MISEV Checklist Section 4b. MISEV considers one or more of characterization assays to be mandatory: detection of characteristic EV-embedded proteins like tetraspanins or EV bound proteins like Alix, determination of contaminant protein levels like BSA, and detection of canonical proteins from non-plasma membrane or non-endosome compartments. MISEV also encourages investigators to assess the topology of EV cargo material; for example, what fraction of the nucleic acid components (RNA and DNA) bound extra-luminally, and what amount is packaged within the vesicles’ lipid bilayers?
| 1. Global Characterization (Section 4-b, Table 3) – At least one of the three mandatory assays below | ||||
| Importance | Description | Instrumentation | Rooster Assay | |
| +++ Mandatory | Transmembrane or GPI-anchored protein localized in cells at plasma membrane or endosomes |  |
ProteinSimple Jess
Fully Automated Western Blots – to detect the presence of protein and amount |
CD9, CD81, CD63 to indicate plasma membrane or endosome origin |
| +++ Mandatory | Cytosolic protein with membrane-binding or – association capacity |  |
ProteinSimple Jess
Fully Automated Western Blots – to detect the presence of protein and amount |
Alix, TSG101 to indicate the endosomal pathway |
| +++ Mandatory | Assessment of presence/absence of expected contaminants |  |
Biotek Syn2 20
Plate Reader – absorbance, luminescence, and fluorescence for protein quantification |
Albumin Contaminant Quantification |
| ++ Mandatory if applicable | Presence of proteins associated with compartments other than plasma membrane or endosomes |  |
ProteinSimple Jess
Fully Automated Western Blots – to detect the presence of protein |
Custom Assay |
 |
Biotek Syn2 20
Plate Reader – absorbance, luminescence, and fluorescence for protein quantification |
Custom Assay | ||
| ++ Mandatory if applicable | Presence of soluble secreted proteins and their likely transmembrane ligands |  |
Biotek Syn2 20
Plate Reader – absorbance, luminescence, and fluorescence for protein quantification |
Custom ELISA |
| + Encouraged
|
Topology of the relevant functional components (Section 4-d) |  |
Agilent
|
Total RNA/miRNA |
 |
QuantStudio 5
|
Specific miRNA Expression | ||
 |
Biotek Syn2 20
Plate Reader – absorbance, luminescence, and fluorescence for protein quantification |
Picogreen Assay to quantify DNA | ||
The MISEV Checklist Section 4c. MISEV recommends as mandatory “Images of single EVs by wide-field and close-up: e.g. electron microscopy, scanning probe microscopy, super-resolution fluorescence microscopy.” These advanced techniques to obtain detailed morphological information demand rare expertise and obtained only in low throughput. RoosterBio is thus not equipped to perform these at present time but is able to recommend excellent external technical services. This section of the checklist also recommends “Non-image-based method analysing large numbers of single EVs: NTA, TRPS, FCS, high-resolution flow cytometry, multi-angle light-scattering, Raman spectroscopy, etc.” RoosterBio is able to provide for analytics here with its NanoFCM that can dissect individual EV vesicle surface markers and the NanoSight for its NTA-based particle size data.
| 3. Single EV Characterization (Section 4-c) | ||||
| Importance | Description | Instrumentation | Rooster Assay | |
| +++ Mandatory | Images of single EVs by wide-field and close-up: e.g. electron microscopy, scanning probe microscopy, super-resolution fluorescence microscopy | NA | NA | NA |
| +++ Mandatory | Non-image-based method analysing large numbers of single EVs: NTA, TRPS, FCS, high-resolution flow cytometry, multi-angle light-scattering, Raman spectroscopy, etc. |  |
NanoSight 18
Nano Tracking Analysis – assess particle count and size distribution |
EV Concentration in particles/mL and EV diameter distribution |
 |
NanoFCM
Nanoparticle Flow Cytometry – counts, particle size, purity, surface markers and specific cargo detection |
Custom Assay | ||
The MISEV 2018 position statement concludes with:
“Finally, there are exceptions to every rule. MISEV 2018 is meant to guide and improve the field, not stifle it. If MISEV recommendations and requirements cannot be met, authors will then need to explain their unique situation and describe their attempts to meet the guidelines and the reason for failure. These guidelines will also continue to evolve.”
The 2022 draft Document also adds that the MISEV position is not intended to be “a one-size-fits-all blueprint or a substitute for common sense” or among other things, a “barrier to innovation.” 14 That’s plainly obvious based on the unfettered explosion in recent related publications and patent applications, many of which look to MISEV for guidance. Accordingly, RoosterBio has proudly attended recent ISEV annual meetings where lively discussions and debate continue on EV/exosome analytics,22, 23, 24, 25 and we’re thrilled to present data at 2023’s Annual Meeting in Seattle. 26 We’ve certainly observed “punctuated equilibrium” as this exciting study rapidly radiates its own evolutionary variants in so many directions across biotechnology’s landscape. Looking back on this blog’s analogy of the wing design, we recall that evolution has “innovated” wings multiple times in animal life across the natural world with insects, birds, bats, and even fish; wherever there’s a niche, life tends to fill it—even in apparent defiance of gravity! With RoosterBio’s newly built capabilities in EV analytical services, we’re here to assist exosome product developers develop and exercise the most optimized “wings” of their own based via the template offered by MISEV.
References
- Thery, C.; and Witwer, K. W., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 2018, 7 (1), 1535750. 10.1080/20013078.2018.1535750
- Willms, E.; Cabanas, C.; Mager, I.; Wood, M. J. A.; Vader, P. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front Immunol 2018, 9, 738. 10.3389/fimmu.2018.00738
- Ginini, L.; Billan, S.; Fridman, E.; Gil, Z. Insight into Extracellular Vesicle-Cell Communication: From Cell Recognition to Intracellular Fate. Cells 2022, 11 (9). 10.3390/cells11091375
- “Where Do We Come From? What Are We? Where Are We Going?” Know Your Process & Define Your Product with EV/Exosome Analytics. https://www.roosterbio.com/blog/where-do-we-come-from-what-are-we-where-are-we-going-know-your-process-define-your-product-with-ev-exosome-analytics/.
- Adlerz, K. and Patel, D. MSC-EVs Emerge as Clinical Therapies. https://www.roosterbio.com/blog/msc-evs-emerge-as-clinical-therapies/.
- Lenzini, S. Big Effects in Small Packages: What Are Extracellular Vesicles, Exosomes, & Microvesicles & Why Are They En Route to the Clinic? https://www.roosterbio.com/blog/big-effects-in-small-packages-what-are-extracellular-vesicles-exosomes-microvesicles-why-are-they-en-route-to-the-clinic/.
- Moller, A.; Lobb, R. J. The evolving translational potential of small extracellular vesicles in cancer. Nat Rev Cancer 2020, 20 (12), 697-709. 10.1038/s41568-020-00299-w
- Grimaldi, A. M.; Salvatore, M.; Cavaliere, C. Diagnostic and prognostic significance of extracellular vesicles in prostate cancer drug resistance: A systematic review of the literature. Prostate Cancer Prostatic Dis 2022. 10.1038/s41391-022-00521-w
- Nicoletti, A.; Negri, M.; Paratore, M.; Vitale, F.; Ainora, M. E.; Nista, E. C.; Gasbarrini, A.; Zocco, M. A.; Zileri Dal Verme, L. Diagnostic and Prognostic Role of Extracellular Vesicles in Pancreatic Cancer: Current Evidence and Future Perspectives. Int J Mol Sci 2023, 24 (1). 10.3390/ijms24010885
- Burrello, J.; Monticone, S.; Gai, C.; Gomez, Y.; Kholia, S.; Camussi, G. Stem Cell-Derived Extracellular Vesicles and Immune-Modulation. Front Cell Dev Biol 2016, 4, 83. 10.3389/fcell.2016.00083
- Liu, Y.; Holmes, C. Tissue Regeneration Capacity of Extracellular Vesicles Isolated From Bone Marrow-Derived and Adipose-Derived Mesenchymal Stromal/Stem Cells. Front Cell Dev Biol 2021, 9, 648098. 10.3389/fcell.2021.648098
- Kogure, A.; Yoshioka, Y.; Ochiya, T. Extracellular Vesicles in Cancer Metastasis: Potential as Therapeutic Targets and Materials. Int J Mol Sci 2020, 21 (12). 10.3390/ijms21124463
- Lotvall, J.; Hill, A. F.; Hochberg, F.; Buzas, E. I.; Di Vizio, D.; Gardiner, C.; Gho, Y. S.; Kurochkin, I. V.; Mathivanan, S.; Quesenberry, P.; Sahoo, S.; Tahara, H.; Wauben, M. H.; Witwer, K. W.; Thery, C. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles 2014, 3, 26913. 10.3402/jev.v3.26913
- Revision Committee, MISEV. Minimal information for studies of extracellular vesicles 2022 (MISEV2022): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2018 guidelines. https://isev.memberclicks.net/assets/docs/MISEV22%20Draft.pdf.
- Couch, Y.; Buzas, E. I.; Di Vizio, D.; Gho, Y. S.; Harrison, P.; Hill, A. F.; Lotvall, J.; Raposo, G.; Stahl, P. D.; Thery, C.; Witwer, K. W.; Carter, D. R. F. A brief history of nearly EV-erything – The rise and rise of extracellular vesicles. J Extracell Vesicles 2021, 10 (14), e12144. 10.1002/jev2.12144
- Doyle, L. M.; Wang, M. Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8 (7). 10.3390/cells8070727
- chemometec.com. NucleoCounting – Count cells intelligently with the Via1-Cassette™ and NucleoCounter® NC-200™. https://chemometec.com/wp-content/uploads/2021/03/NucleoCounter-NC-200.pdf.
- malvernpanalytical.com. NanoSight NS300 – High resolution nanoparticle size and concentration data. https://www.malvernpanalytical.com/en/products/product-range/nanosight-range/nanosight-ns300.
- particle-metrix.com. ZetaView® QUATT – Technical Data. https://particle-metrix.com/wp-content/uploads/2022/05/Datasheet-ZetaView
- BioTek. Synergy™2 – Operator’s Manual. https://www.pedsresearch.org/uploads/lab/doc/Synergy_2.pdf.
- agilent.com. 2100 Bioanalyzer Instrument. https://www.agilent.com/en/product/automated-electrophoresis/bioanalyzer-systems/bioanalyzer-instrument/2100-bioanalyzer-instrument-228250.
- Carson, J. and Lembong, J. ISEV2020 Summary – Part I: To Harness Heterogeneity, Extracellular Vesicle (EV) Differences Explored with Deference. https://www.roosterbio.com/blog/isev-2020-summary-part-1/.
- Lembong, J., Carson, J., and Rowley, J. ISEV2020 Summary – Part II: Whether Upstream or Downstream, EV Manufacturing is Becoming Seaworthy in 2020. https://www.roosterbio.com/blog/isev-2020-summary-part-2/.
- RoosterBio Joins the Action at ISCT, ASGCT, & ISEV in May 2022. https://www.roosterbio.com/blog/roosterbio-joins-the-action-at-isct-asgct-isev-in-may-2022/.
- Lenzini, S. Highlights from ISEV 2022: In-Person Inspires In-Depth EV/Exosome Discussions & Analysis. https://www.roosterbio.com/blog/highlights-from-isev-2022-in-person-inspires-in-depth-ev-exosome-discussions-analysis/.
- Upstream, Downstream, & In Between: RoosterBio Collaborations Featured at May 2023 Conferences (ISEV, ISCT, & ASGCT). https://www.roosterbio.com/blog/upstream-downstream-in-between-roosterbio-collaborations-featured-at-may-2023-conferences-isev-isct-asgct/.
