MSC-EV Signatures — Not Quite Signed, Sealed, & Delivered
Extracellular vesicles (or EVs, sometimes called “exosomes”) have been investigated for at least two decades 1, 2, 3 due to their prominence in normal human development, 4 disease, 5, 6 diagnosis, 7, 8 and now, next-generation advanced therapies.9, 10, 11, 12, 13, 14, 15, 16, 17, 18 Regarding the latter, mesenchymal stromal/stem cells (MSCs) have advanced to the leading edge of cellular bioproduction sources of extracellular vesicles for human clinical trials.19, 20 For this reason, preclinical investigators are avidly prospecting for definitive intraluminal molecular components of MSC-EVs to use as critical quality attributes (CQAs).21
Nevertheless, key molecular determinants of extracellular vesicle therapeutic relevance remain elusive. There’s abundant evidence to suggest that ligands on or associated with the external surfaces of extracellular vesicles contribute to much of their biofunction.22, 23 These interactions promulgate “outside-in” signal transduction cascaded via receptors, kinases, and second messengers on target cells. There’s also evidence that RNAs associate with MSC-EVs, likely existing both external to the EV surface and within the EV membrane. A predominant hypothesis is that, in general, nucleic acid cargoes can be delivered by extracellular vesicles to elicit relevant functions in recipient cells.
Perplexity on the Edge of Complexity?
Valadi, et al’s (2007) landmark publication 24 first demonstrated that RNAse-resistant RNA types (including intact mRNA and micro (mi)RNAs) were packaged in the small extracellular vesicles of live cells.25 When these mouse EVs were transferred to human target cells with “exosomal shuttle RNA” (esRNA), new mouse proteins were generated in the human cells.
Since these results were reported, increased analytical sophistication brought to bear on heterogeneous extracellular vesicle types containing (or associated with) different molecular RNA species, including mRNA, miRNA, tRNA, ribosomal RNA (rRNA), Y-RNA, long non-coding RNA (lncRNA), among others.23, 26, 27, 28, 29, 30, 31 These descriptive studies raise several questions, though. For example, extracellular vesicle biogenesis routes include formation out of the plasma membrane (microvesicles, ARRMs, Arc/Gag retrotransposon products), or through multivesicular bodies (i.e., exosomes). EV cargoes can vary according to their mode of biogenesis and their exposures to different environments and stimuli. This means that EV populations can be vastly heterogeneous and therefore, different EV populations within the same sample likely include different RNA cargoes.
Endogenous expression of various RNA types has been correlated with extracellular vesicle function. Moreover, overexpression or exogenous loading of RNAs into extracellular vesicles has been shown to elicit functional effects in certain contexts. 32, 33 Studies that knockdown miRNA regulatory elements such as Dicer have attributed EV function to miRNA cargoes. 34 However, it remains generally unclear to what extent the naturally occurring levels of RNA species within EVs are functionally relevant. Low levels of RNAs in extracellular samples are notoriously difficult to detect, and thus conducting the necessary studies to answer these questions requires the ability to produce consistent, large-scale batches of MSC-EVs for analytical testing. From there, these batches can be used to identify RNA species that are detectable and consistently expressed within extracellular vesicles across a range of different MSC tissue types and donors.
RoosterBio Glimpses Into a Clear miRNAome
With potential caveats in mind, we at RoosterBio remain completely undeterred—and very fascinated—by the mysterious RNA cargos within extracellular vesicles that our own team, customers, and partners produce. Why? Specific RNA presence and quantity via qRT-PCR could be compiled into a routine molecular signature to validate consistent hMSC phenotype and EV cargo integrity across cell donors, tissue origins, and preparation conditions.
As a starting point, we chose to profile miRNAs in hMSC-EVs, since a wealth of literature exists for miRNA contents within hMSC-EVs specifically. Using the TaqMan™ Advanced miRNA Human A and B 96-well Plates, we probed for 792 unique miRNAs via RNA preparations extracted from EVs prepared from one bone marrow hMSC donor. From this initial screen, we determined that some miRNAs were more abundant than others, including some species with noted therapeutic effects of interest. This panel included miR-125b-5p (anti-inflammation),35 miR-23a-3p (angiogenesis),32 miR-221-3p (cardioprotection),36 miR-598-3p (neurological),37 miR-92a-3p (anti-apoptotic signaling and brain recovery),38 and miR-16a-5p (involvement in p53 signaling).39, 40
From here, we recognized that miR-16a-5p has been used previously as a reference miRNA since it is stably expressed in a variety of cell types. 39, 41 A successful quantitative polymerase chain reaction (qPCR) assay to detect presence of RNA in samples includes: (1) a negative control to ensure that RNA is not detected in samples lacking RNA, and (2) a reference standard that is consistently expressed across tested samples.
To develop this assay, we prepared hMSC-EV samples from MSCs derived from two different tissue types (bone marrow [BM] and umbilical cord [UC]) and two different donors (donor 1 and donor 2). Conditioned medium samples were first clarified by centrifugation to remove cell debris (500g x 5 minutes), followed by ultracentrifugation to concentrate EV material. Small RNA was then extracted from the EV-containing pellet using the Qiagen miRNeasy kit system. RNAs were then reverse transcribed to cDNA for use in qPCR, where nucleic acid concentrations were modified to be equivalent across samples (400 pg/mL) for the assay. Using the TaqMan MicroRNA (ThermoFisher) reagents against the miRNA targets identified above, the no-template control results confirm the lack of non-specific detection for all tested miRNA species (Figure 1). Furthermore, quantification of the reference standard miR-16a-5p showed remarkably consistent results for all samples, confirming its relevance as a miRNA reference standard.
After developing this qPCR assay, we then evaluated expression of the above miRNA panel using the same samples tested in Fig. 1. Results showed the detection of all tested miRNA species across all tested samples including different tissue types (BM and UC) and different donors (donor 1 and donor 2) (Figure 2). Normalization of Ct values to those of the reference standard miR-16a-5p (Figure 3) shows a clear pattern of expression which is relatively consistent across tissue types and donors.
A Few Reflections
In summary, we conducted initial studies to identify RNA species present in hMSC-EVs. First, we screened 792 unique miRNA species and identified five abundant targets including miR-16a-5p, a species used previously as a miRNA reference standard for data normalization. Next, we developed a qPCR assay to quantify the presence of RNA species in hMSC-EV samples, using miR-16a-5p as a reference standard. Finally, we prepared MSC-EVs from two different tissue types (BM and UC) and two different donors (donor 1 and donor 2) and showed consistent expression of miRNA targets. Therefore, this assay can be used to validate the consistent presence of targeted RNA species across hMSC-EVs prepared from different samples.
These results represent a first step towards identifying RNA species in hMSC-EVs that are relevant for therapeutic purposes, and this approach serves as a framework for potential CQA identification. From this identification step, targets will require more extensive functional experiments to validate their relevance as CQAs. Here, we identified a small group of distinct miRNA species. Further work will test additional miRNAs as well as expand to other RNA types and different species within those types.
As RoosterBio penetrates the identification of CQAs for MSC-EVs, we invite you to join us as your partner in extracellular vesicle analytics assays. We’ve already developed an extensive panel of standard metrics and methods to assist your process development and manufacturing of extracellular vesicles/exosomes, and are always seeking additional ones of benefit this exciting field of research.
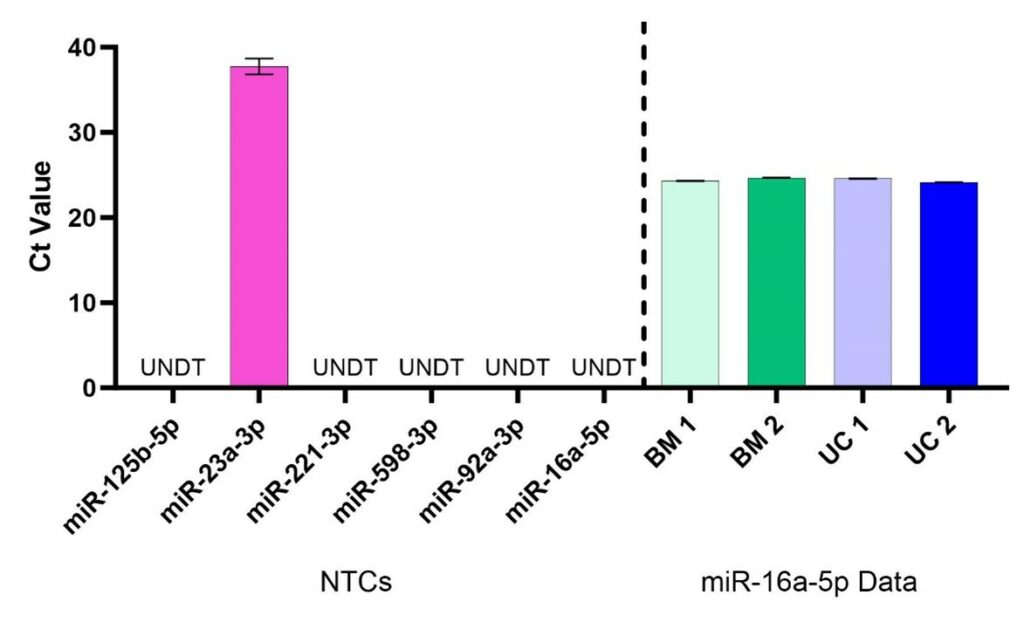
Figure 1. No-template control (NTC, left of dotted line) and reference miRNA (right of dotted line) critical threshold (Ct) values. For NTC samples, all targets except one (that for miR-23a-3p) showed Undetermined (UNDT) Ct values, meaning that amplification of cDNA was not detected. Ct > 35 are associated with extremely low nucleic acid content and can be considered assay noise. Ct values for miR-16a-5p were < 25, indicating strong detectability, and consistent with low variability across all samples (BM 1 %CV = 0.17%, BM 2 %CV = 0.18%, UC 1 %CV = 0.13%, UC 2 %CV = 0.13%). N=4, error bars are standard deviation.
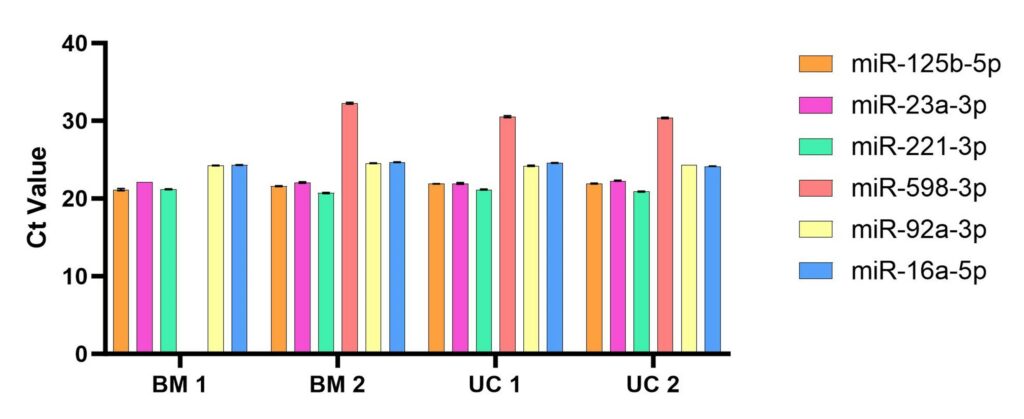
Figure 2. Ct Values for qRT-PCR runs across the six probes for miR-125b-5p, -23a-3p, 221-3p, -598-3p, -92a-3p, and -16a-5p (reference). Lower Ct denotes a smaller number of PCR cycles sufficient for detectable amplification, i.e., the Ct is inversely proportional to miRNA abundance. BM 1 and BM 2 are samples derived from extracellular vesicles collected from bone marrow hMSCs UC 1 and UC 2 are from umbilical cord hMSC-EVs. For BM 1, miR-598-3p did not reach a critical threshold, indicating undetectable levels of this RNA species in this sample. N=4, error bars are standard deviation.
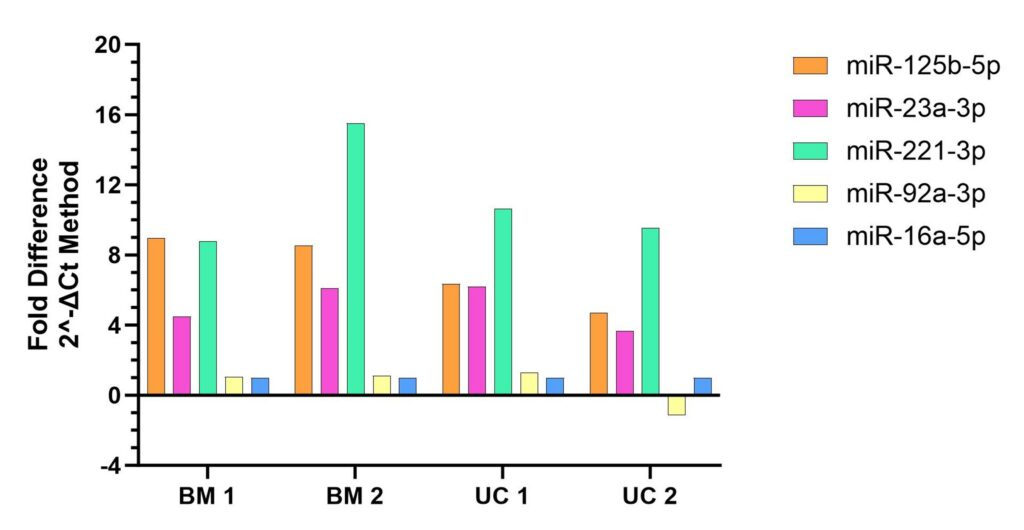
Figure 3. Reconfigured data from Fig.2, expressed as a fold difference relative to the reference miRNA (miR-16a-5-p).
References
- Thery, C.; Zitvogel, L.; Amigorena, S., Exosomes: composition, biogenesis and function. Nat Rev Immunol 2002, 2 (8), 569-79. 10.1038/nri855
- Couch, Y.; Buzas, E. I.; Di Vizio, D.; Gho, Y. S.; Harrison, P.; Hill, A. F.; Lotvall, J.; Raposo, G.; Stahl, P. D.; Thery, C.; Witwer, K. W.; Carter, D. R. F., A brief history of nearly EV-erything – The rise and rise of extracellular vesicles. J Extracell Vesicles 2021, 10 (14), e12144. 10.1002/jev2.12144
- Kalluri, R.; LeBleu, V. S., The biology, function, and biomedical applications of exosomes. Science 2020, 367 (6478). 10.1126/science.aau6977
- Tannetta, D.; Dragovic, R.; Alyahyaei, Z.; Southcombe, J., Extracellular vesicles and reproduction-promotion of successful pregnancy. Cell Mol Immunol 2014, 11 (6), 548-63. 10.1038/cmi.2014.42
- Wortzel, I.; Dror, S.; Kenific, C. M.; Lyden, D., Exosome-Mediated Metastasis: Communication from a Distance. Dev Cell 2019, 49 (3), 347-360. 10.1016/j.devcel.2019.04.011
- Hill, A. F., Extracellular Vesicles and Neurodegenerative Diseases. J Neurosci 2019, 39 (47), 9269-9273. 10.1523/JNEUROSCI.0147-18.2019
- Yu, D.; Li, Y.; Wang, M.; Gu, J.; Xu, W.; Cai, H.; Fang, X.; Zhang, X., Exosomes as a new frontier of cancer liquid biopsy. Mol Cancer 2022, 21 (1), 56. 10.1186/s12943-022-01509-9
- Yu, X.; Odenthal, M.; Fries, J. W., Exosomes as miRNA Carriers: Formation-Function-Future. Int J Mol Sci 2016, 17 (12). 10.3390/ijms17122028
- Kim, S. H.; Bianco, N.; Menon, R.; Lechman, E. R.; Shufesky, W. J.; Morelli, A. E.; Robbins, P. D., Exosomes derived from genetically modified DC expressing FasL are anti-inflammatory and immunosuppressive. Mol Ther 2006, 13 (2), 289-300. 10.1016/j.ymthe.2005.09.015
- Zeelenberg, I. S.; Ostrowski, M.; Krumeich, S.; Bobrie, A.; Jancic, C.; Boissonnas, A.; Delcayre, A.; Le Pecq, J. B.; Combadiere, B.; Amigorena, S.; Thery, C., Targeting tumor antigens to secreted membrane vesicles in vivo induces efficient antitumor immune responses. Cancer Res 2008, 68 (4), 1228-35. 10.1158/0008-5472.CAN-07-3163
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M. J., Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol 2011, 29 (4), 341-5. 10.1038/nbt.1807
- Wahlgren, J.; Statello, L.; Skogberg, G.; Telemo, E.; Valadi, H., Delivery of Small Interfering RNAs to Cells via Exosomes. Methods Mol Biol 2016, 1364, 105-25. 10.1007/978-1-4939-3112-5_10
- Stranford, D. M.; Leonard, J. N., Delivery of Biomolecules via Extracellular Vesicles: A Budding Therapeutic Strategy. Adv Genet 2017, 98, 155-175. 10.1016/bs.adgen.2017.08.002
- Izco, M.; Blesa, J.; Schleef, M.; Schmeer, M.; Porcari, R.; Al-Shawi, R.; Ellmerich, S.; de Toro, M.; Gardiner, C.; Seow, Y.; Reinares-Sebastian, A.; Forcen, R.; Simons, J. P.; Bellotti, V.; Cooper, J. M.; Alvarez-Erviti, L., Systemic Exosomal Delivery of shRNA Minicircles Prevents Parkinsonian Pathology. Mol Ther 2019, 27 (12), 2111-2122. 10.1016/j.ymthe.2019.08.010
- Kooijmans, S. A. A.; de Jong, O. G.; Schiffelers, R. M., Exploring interactions between extracellular vesicles and cells for innovative drug delivery system design. Adv Drug Deliv Rev 2021, 173, 252-278. 10.1016/j.addr.2021.03.017
- Gupta, D.; Wiklander, O. P. B.; Gorgens, A.; Conceicao, M.; Corso, G.; Liang, X.; Seow, Y.; Balusu, S.; Feldin, U.; Bostancioglu, B.; Jawad, R.; Mamand, D. R.; Lee, Y. X. F.; Hean, J.; Mager, I.; Roberts, T. C.; Gustafsson, M.; Mohammad, D. K.; Sork, H.; Backlund, A.; Lundin, P.; de Fougerolles, A.; Smith, C. I. E.; Wood, M. J. A.; Vandenbroucke, R. E.; Nordin, J. Z.; El-Andaloussi, S., Amelioration of systemic inflammation via the display of two different decoy protein receptors on extracellular vesicles. Nat Biomed Eng 2021, 5 (9), 1084-1098. 10.1038/s41551-021-00792-z
- Mitrut, R. E.; Stranford, D. M.; Chan, J. M.; Bailey, M. D.; Luo, M.; Meade, T. J.; Wang, M.; Leonard, J. N., HaloTag display enables quantitative single-particle characterization and functionalization of engineered extracellular vesicles. bioRxiv 2023. 10.1101/2023.09.25.559433
- Liu, Y.; Li, D.; Liu, Z.; Zhou, Y.; Chu, D.; Li, X.; Jiang, X.; Hou, D.; Chen, X.; Chen, Y.; Yang, Z.; Jin, L.; Jiang, W.; Tian, C.; Zhou, G.; Zen, K.; Zhang, J.; Zhang, Y.; Li, J.; Zhang, C. Y., Targeted exosome-mediated delivery of opioid receptor Mu siRNA for the treatment of morphine relapse. Sci Rep 2015, 5, 17543. 10.1038/srep17543
- Toh, W. S.; Yarani, R.; El Andaloussi, S.; Cho, B. S.; Choi, C.; Corteling, R.; De Fougerolles, A.; Gimona, M.; Herz, J.; Khoury, M.; Robbins, P. D.; Williams, D.; Weiss, D. J.; Rohde, E.; Giebel, B.; Lim, S. K., A report on the International Society for Cell & Gene Therapy 2022 Scientific Signature Series, “Therapeutic advances with native and engineered human extracellular vesicles”. Cytotherapy 2023, 25 (8), 810-814. 10.1016/j.jcyt.2023.02.009
- Yarani, R.; Lim, S. K.; Giebel, B., Mesenchymal stromal cells extracellular vesicles; unlocking the potential. Cytotherapy 2023, 25 (8), 808-809. 10.1016/j.jcyt.2023.05.008
- Asgarpour, K.; Shojaei, Z.; Amiri, F.; Ai, J.; Mahjoubin-Tehran, M.; Ghasemi, F.; ArefNezhad, R.; Hamblin, M. R.; Mirzaei, H., Exosomal microRNAs derived from mesenchymal stem cells: cell-to-cell messages. Cell Commun Signal 2020, 18 (1), 149. 10.1186/s12964-020-00650-6
- Toh, W. S.; Lai, R. C.; Zhang, B.; Lim, S. K., MSC exosome works through a protein-based mechanism of action. Biochem Soc Trans 2018, 46 (4), 843-853. 10.1042/BST20180079
- Jeppesen, D. K.; Fenix, A. M.; Franklin, J. L.; Higginbotham, J. N.; Zhang, Q.; Zimmerman, L. J.; Liebler, D. C.; Ping, J.; Liu, Q.; Evans, R.; Fissell, W. H.; Patton, J. G.; Rome, L. H.; Burnette, D. T.; Coffey, R. J., Reassessment of Exosome Composition. Cell 2019, 177 (2), 428-445 e18. 10.1016/j.cell.2019.02.029
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J. J.; Lötvall, J. O., Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007, 9 (6), 654-9. 10.1038/ncb1596
- Lotvall, J.; Valadi, H., Cell to cell signalling via exosomes through esRNA. Cell Adh Migr 2007, 1 (3), 156-8. 10.4161/cam.1.3.5114
- Mateescu, B.; Kowal, E. J.; van Balkom, B. W.; Bartel, S.; Bhattacharyya, S. N.; Buzas, E. I.; Buck, A. H.; de Candia, P.; Chow, F. W.; Das, S.; Driedonks, T. A.; Fernandez-Messina, L.; Haderk, F.; Hill, A. F.; Jones, J. C.; Van Keuren-Jensen, K. R.; Lai, C. P.; Lasser, C.; Liegro, I. D.; Lunavat, T. R.; Lorenowicz, M. J.; Maas, S. L.; Mager, I.; Mittelbrunn, M.; Momma, S.; Mukherjee, K.; Nawaz, M.; Pegtel, D. M.; Pfaffl, M. W.; Schiffelers, R. M.; Tahara, H.; Thery, C.; Tosar, J. P.; Wauben, M. H.; Witwer, K. W.; Nolte-‘t Hoen, E. N., Obstacles and opportunities in the functional analysis of extracellular vesicle RNA – an ISEV position paper. J Extracell Vesicles 2017, 6 (1), 1286095. 10.1080/20013078.2017.1286095
- Fatima, F.; Ekstrom, K.; Nazarenko, I.; Maugeri, M.; Valadi, H.; Hill, A. F.; Camussi, G.; Nawaz, M., Non-coding RNAs in Mesenchymal Stem Cell-Derived Extracellular Vesicles: Deciphering Regulatory Roles in Stem Cell Potency, Inflammatory Resolve, and Tissue Regeneration. Front Genet 2017, 8, 161. 10.3389/fgene.2017.00161
- Pastuzyn, E. D.; Day, C. E.; Kearns, R. B.; Kyrke-Smith, M.; Taibi, A. V.; McCormick, J.; Yoder, N.; Belnap, D. M.; Erlendsson, S.; Morado, D. R.; Briggs, J. A. G.; Feschotte, C.; Shepherd, J. D., The Neuronal Gene Arc Encodes a Repurposed Retrotransposon Gag Protein that Mediates Intercellular RNA Transfer. Cell 2018, 172 (1-2), 275-288 e18. 10.1016/j.cell.2017.12.024
- Kim, K. M.; Abdelmohsen, K.; Mustapic, M.; Kapogiannis, D.; Gorospe, M., RNA in extracellular vesicles. Wiley Interdiscip Rev RNA 2017, 8 (4). 10.1002/wrna.1413
- Li, M.; Zeringer, E.; Barta, T.; Schageman, J.; Cheng, A.; Vlassov, A. V., Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philosophical Transactions of the Royal Society B: Biological Sciences 2014, 369 (1652), 20130502. doi:10.1098/rstb.2013.0502
- Corrado, C.; Barreca, M. M.; Zichittella, C.; Alessandro, R.; Conigliaro, A., Molecular Mediators of RNA Loading into Extracellular Vesicles. Cells 2021, 10 (12), 3355. 10.3390/cells10123355
- Ferguson, S. W.; Wang, J.; Lee, C. J.; Liu, M.; Neelamegham, S.; Canty, J. M.; Nguyen, J., The microRNA regulatory landscape of MSC-derived exosomes: a systems view. Sci Rep 2018, 8 (1), 1419. 10.1038/s41598-018-19581-x
- Dalmizrak, A.; Dalmizrak, O., Mesenchymal stem cell-derived exosomes as new tools for delivery of miRNAs in the treatment of cancer. Frontiers in Bioengineering and Biotechnology 2022, 10. 10.3389/fbioe.2022.956563
- Shirazi, S.; Huang, C.-C.; Kang, M.; Lu, Y.; Ravindran, S.; Cooper, L. F., The importance of cellular and exosomal miRNAs in mesenchymal stem cell osteoblastic differentiation. Scientific Reports 2021, 11 (1), 5953. 10.1038/s41598-021-85306-2
- Matsukawa, T.; Sakai, T.; Yonezawa, T.; Hiraiwa, H.; Hamada, T.; Nakashima, M.; Ono, Y.; Ishizuka, S.; Nakahara, H.; Lotz, M. K.; Asahara, H.; Ishiguro, N., MicroRNA-125b regulates the expression of aggrecanase-1 (ADAMTS-4) in human osteoarthritic chondrocytes. Arthritis Res Ther 2013, 15 (1), R28. 10.1186/ar4164
- Sun, L.; Zhu, W.; Zhao, P.; Zhang, J.; Lu, Y.; Zhu, Y.; Zhao, W.; Liu, Y.; Chen, Q.; Zhang, F., Down-Regulated Exosomal MicroRNA-221 – 3p Derived From Senescent Mesenchymal Stem Cells Impairs Heart Repair. Front Cell Dev Biol 2020, 8, 263. 10.3389/fcell.2020.00263
- Zhu, Z.; Huang, X.; Du, M.; Wu, C.; Fu, J.; Tan, W.; Wu, B.; Zhang, J.; Liao, Z. B., Recent advances in the role of miRNAs in post-traumatic stress disorder and traumatic brain injury. Mol Psychiatry 2023, 28 (7), 2630-2644. 10.1038/s41380-023-02126-8
- He, S.; Wang, Z.; Li, Y.; Dong, J.; Xiang, D.; Ren, L.; Guo, L.; Shu, J., MicroRNA-92a-3p enhances functional recovery and suppresses apoptosis after spinal cord injury via targeting phosphatase and tensin homolog. Biosci Rep 2020, 40 (5). 10.1042/BSR20192743
- Ragni, E.; Perucca Orfei, C.; De Luca, P.; Colombini, A.; Vigano, M.; Lugano, G.; Bollati, V.; de Girolamo, L., Identification of miRNA Reference Genes in Extracellular Vesicles from Adipose Derived Mesenchymal Stem Cells for Studying Osteoarthritis. Int J Mol Sci 2019, 20 (5). 10.3390/ijms20051108
- Cai, B.; Ma, M.; Chen, B.; Li, Z.; Abdalla, B. A.; Nie, Q.; Zhang, X., MiR-16-5p targets SESN1 to regulate the p53 signaling pathway, affecting myoblast proliferation and apoptosis, and is involved in myoblast differentiation. Cell Death & Disease 2018, 9 (3), 367. 10.1038/s41419-018-0403-6
- Lange, T.; Stracke, S.; Rettig, R.; Lendeckel, U.; Kuhn, J.; Schlüter, R.; Rippe, V.; Endlich, K.; Endlich, N., Identification of miR-16 as an endogenous reference gene for the normalization of urinary exosomal miRNA expression data from CKD patients. PLoS One 2017, 12 (8), e0183435. 10.1371/journal.pone.0183435
